The p53 protein acts as a central hub that receives, integrates and transmits multiple signals, generated during various stress events, to ensure cell and tissue homeostasis. Although the most important activity of p53 is to act as a direct transcription activator
for several hundreds of genes, it is also able to act as a transcription repressor. Moreover, it has several transcription-independent activities that make the investigation of this protein very complex.
The strongest and most undisputed fact about p53 is the high frequency of TP53 gene alterations in human cancer and the tumor suppressor function of p53 has generally been associated with the induction of cell-cycle arrest, apoptosis, or senescence. Recent studies challenge this dogma by providing evidence that all three of these programs are dispensable for the tumor suppressive role of p53.
Click on the links below for further details
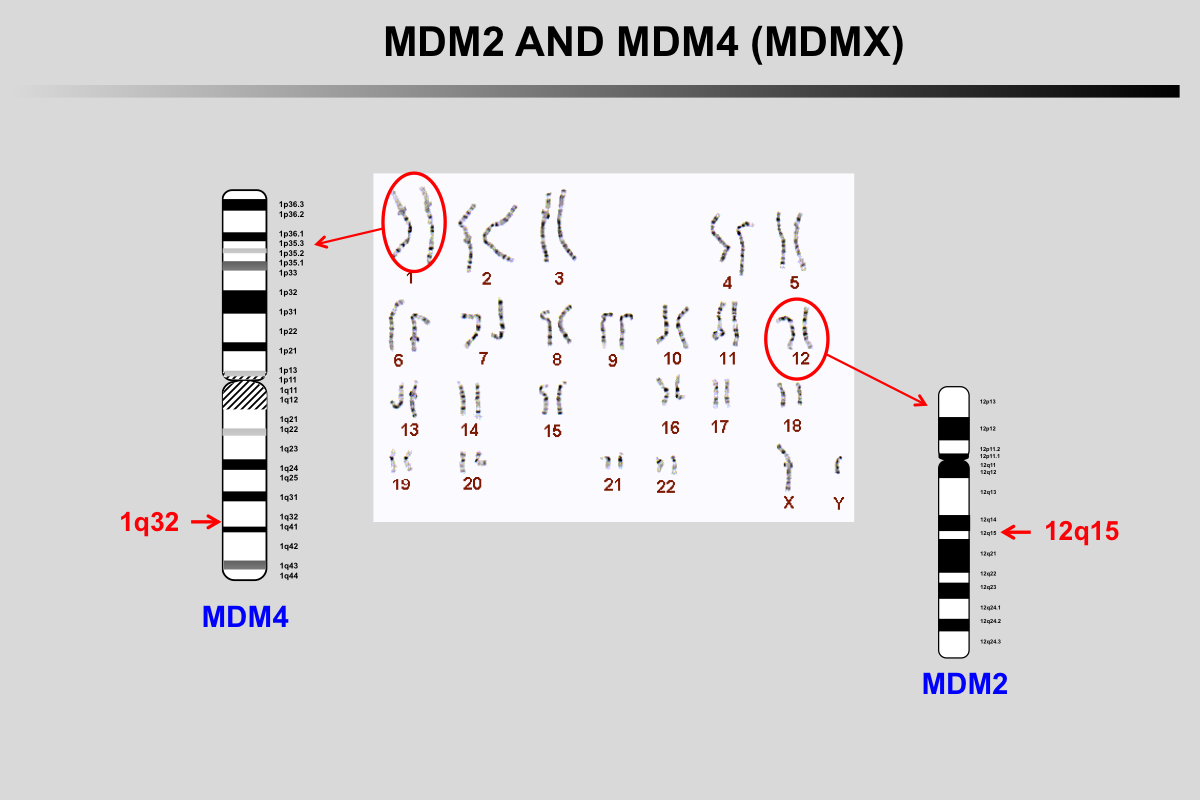
In normal unstressed cells, the level of p53 protein is downregulated by binding of proteins such as MDM2/MDM4, COP1, PIRH2 or JNK that promote p53 degradation via the ubiquitin/proteasome pathway. As most of these genes are upregulated by p53, this leads to a regulation loop that maintains very low p53 levels in normal cells.
Following genotoxic or nongenotoxic stresses, activation of p53 is a two-step process. First, p53 protein level is increased via inhibition of its interaction with mdm2 and other negative regulators. Overtranslation of p53 RNA is also an additional mechanism that also ensures p53 accumulation. Second, a series of modulators (kinases, acetylases) activate p53 transcriptional activity.
A plethora of proteins have been found to bind various regions of p53 in order to regulate the specificity of its activity.
Downstream signaling includes a large series of genes that are activated by the transactivating properties of p53. This occurs via specific DNA binding of the p53 protein to a p53 response element (p53 RE) present in either the promoter or the intron of target genes.
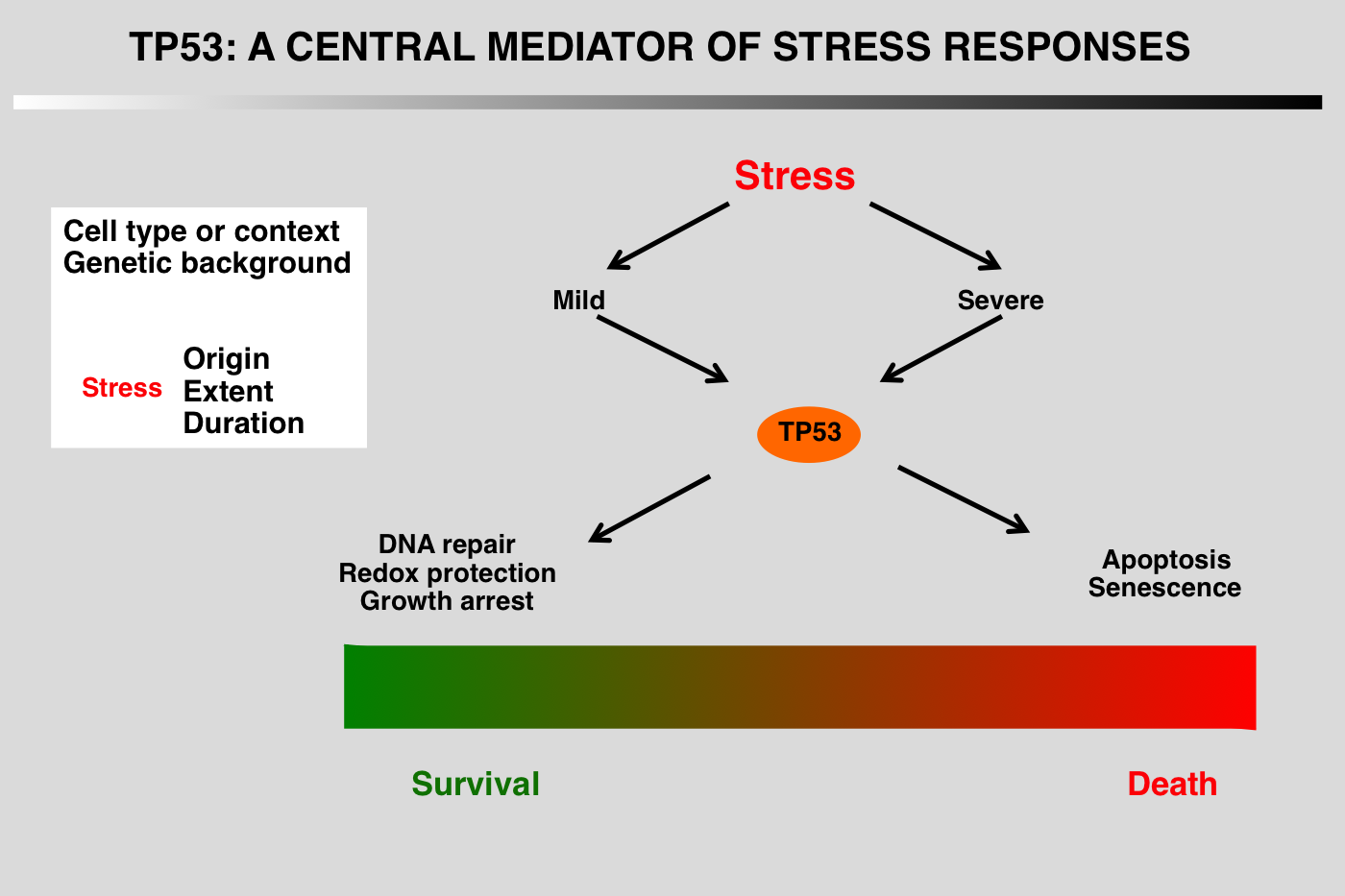
Regardless of the type of stress, the final outcome of p53 activation is either cell cycle arrest and DNA repair or apoptosis, but the mechanism leading to the choice between these fates has not yet been elucidated.
The p53 pathway has been conveniently divided into five parts:
1_Stress signals that activate the pathway
2_Upstream mediators that detect and interpret the upstream signals.
3_Core regulation of p53 via its interaction with several proteins that modulate its stability
4_Downstream events, mainly transcriptional activation or protein-protein interactions
5_Final outcome, growth arrest, apoptosis or DNA repair.
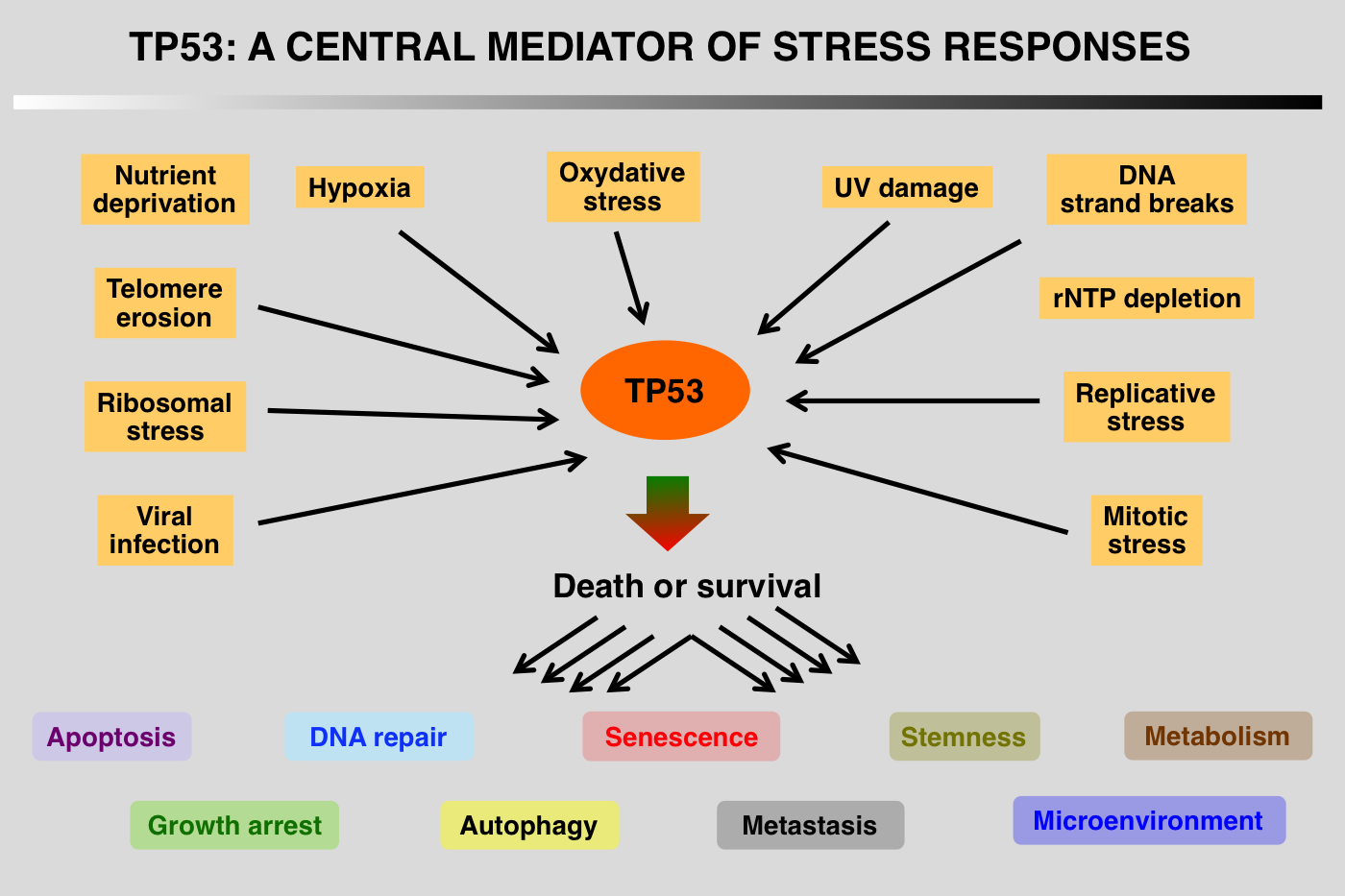
Although the TP53 pathway was initially associated with growth arrest and DNA damage, it is now clear that this protein is part of a central hub that regulates the expression of thousands of genes that control apoptosis, cell cycle arrest, senescence, metabolism, fertility, aging and autophagy. Regulation of p53 turnover is essential to keep p53 activity under control in normal cells. Multiple E3 ligases mediate p53 degradation via the ubiquitination-26S proteasome pathway
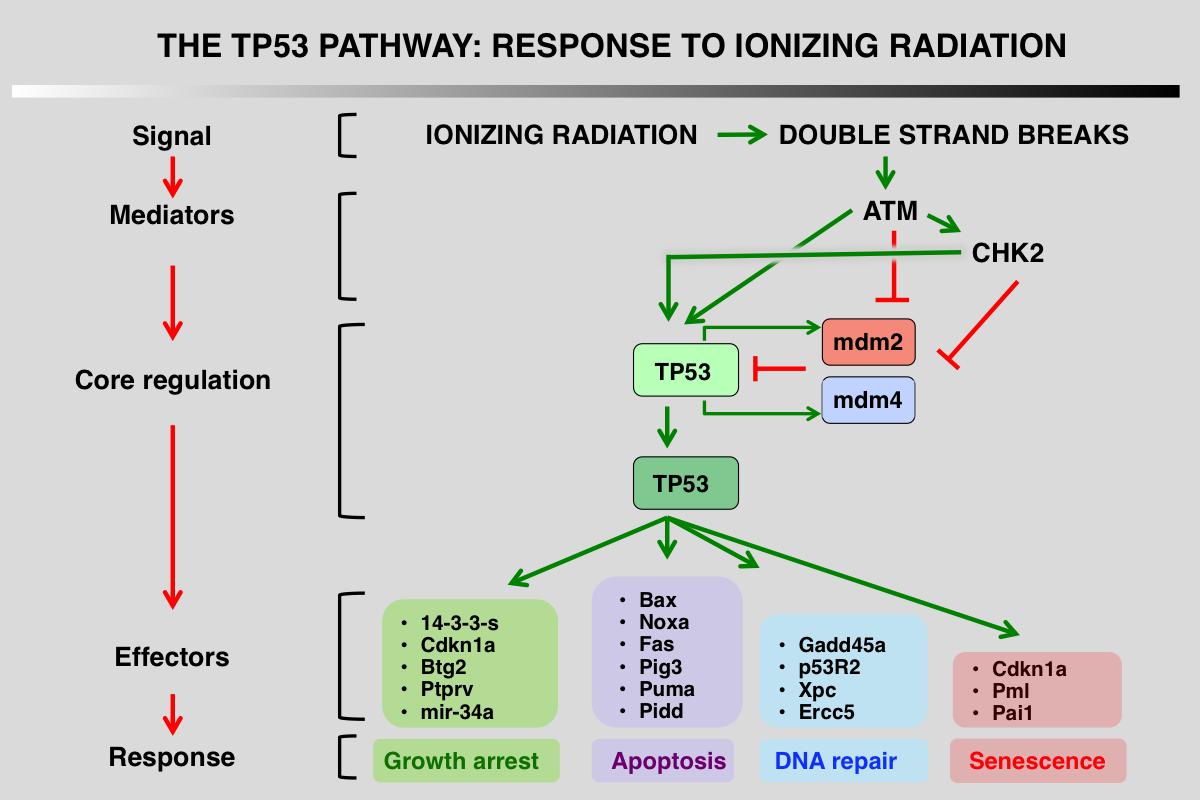
p53 pathway in response to ionizing radiation
1_ Stress signals that activate the pathway: DNA double-strand breaks (DSBs) are highly toxic lesions that can drive genetic instability
2_ Upstream mediators that detect and interpret the upstream signals: ATM and Chk2 kinase phosphorylate p53, mdm2 and mdm4
3_ Core regulation of p53 via its interaction with several proteins that modulate its stability: this interaction is impaired after phosphorylation of the various partners
4_ Downstream events, mainly transcriptional activation or protein-protein interactions: during the DDR response, p21 (CDKN1A) gene is the major target of p53.
5_ Final outcome, growth arrest, apoptosis or DNA repair.
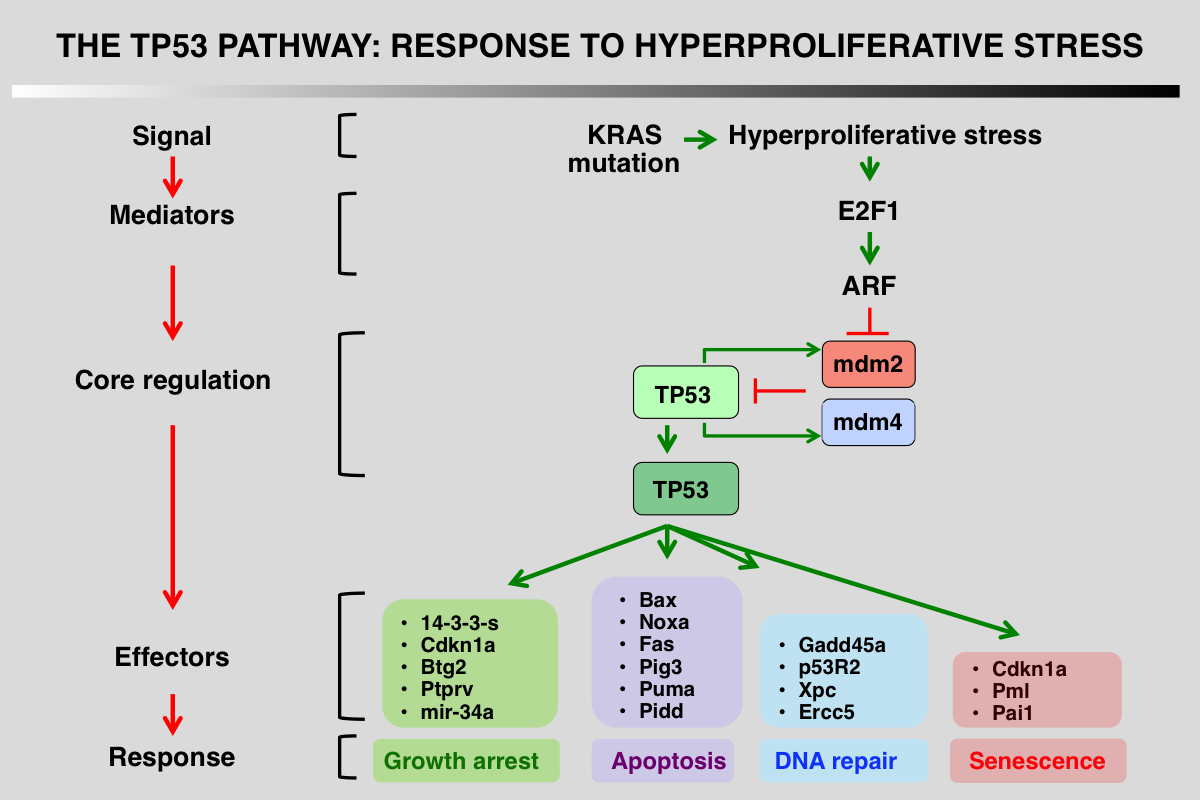
p53 pathway in response to hyperproliferative stress
1_ Stress signals that activate the pathway: KRAS gene mutation.
2_ Upstream mediators that detect and interpret the upstream signals: E2F expression activates transcription of ARF.
3_ Core regulation of p53 via its interaction with several proteins that modulate its stability: ARF binds to mdm2, disrupts p53/mdm2 interaction and sequesters mdm2 in the cell nucleolus.
4_ Downstream events, mainly transcriptional activation or protein-protein interactions: a tissue-specific cell response is induced during this response.
5_ Final outcome, growth arrest, apoptosis or DNA repair.
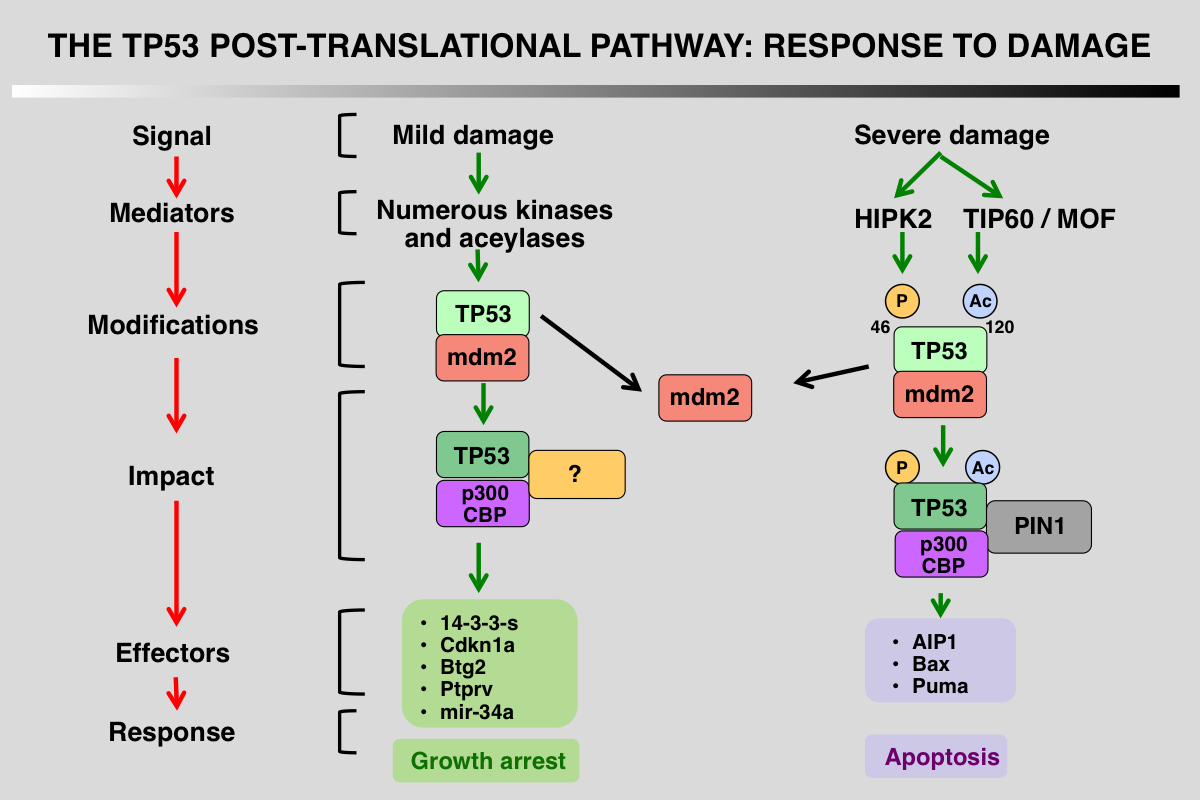
Differential Regulation of p53 phosphorylation after DNA damage
TP53 post-translational modifications are a key feature that orchestrate the p53 response and vary depending on the damage. The first outcome of p53 phosphorylation at the various serines at the amino terminus is recruitment of p300/CBP as well as dissociation of mdm2. Following severe damage, other kinases or acetylases are activated leading to different patterns of modifications and the recruitment of proapoptotic factors.
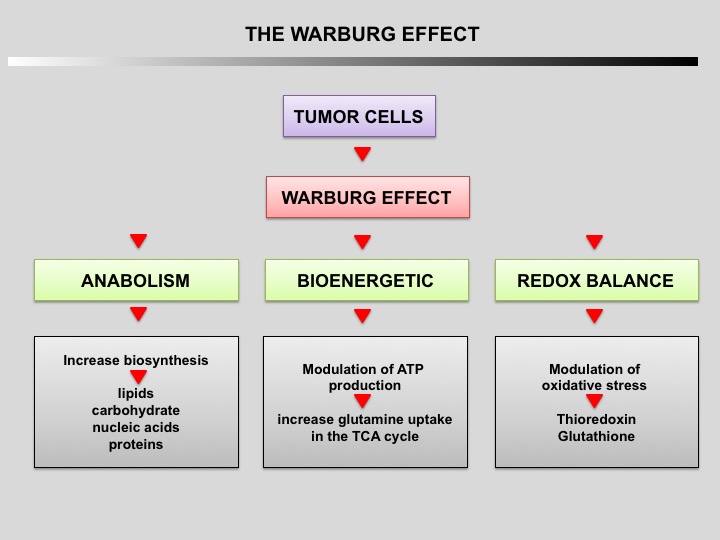
Tumor cells primarily use glycolysis for energy production instead of mitochondrial oxidative phosphorylation like normal cells (Warburg effect discovered by Otto Warburg in 1924). The increase of aerobic glycolysis confers many advantages to cancer cells, including carbon intermediates for biosynthesis, ATP generation, detoxification and support of rapid proliferation
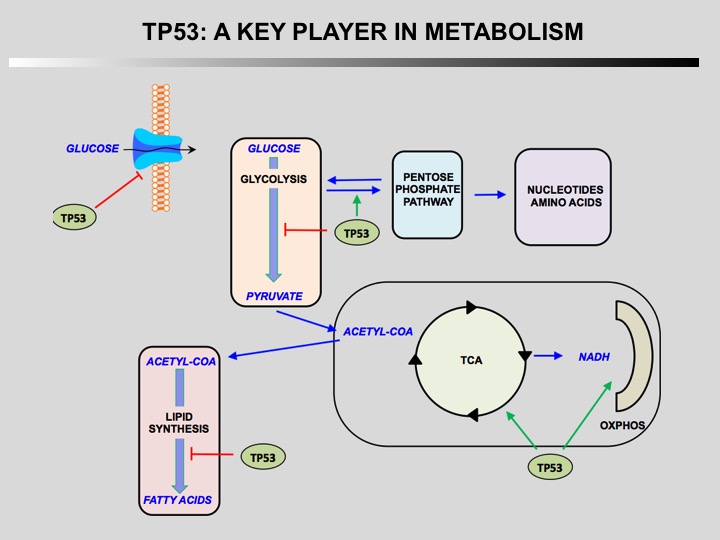
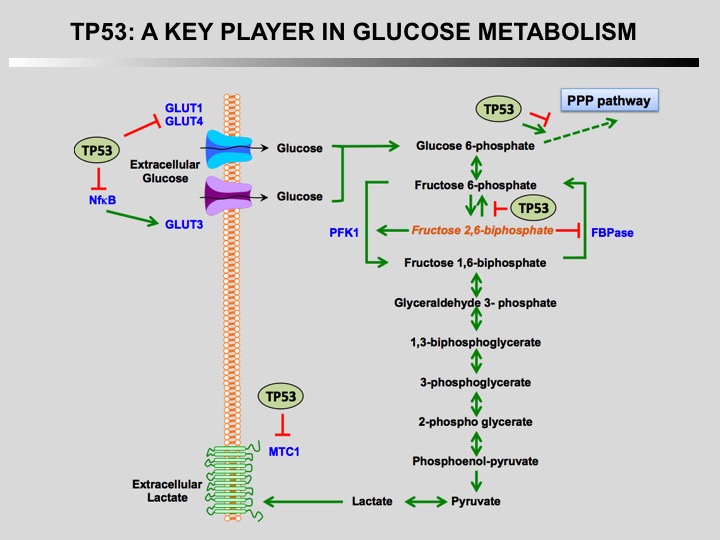
TP53 is a key player in regulation of cellular metabolism. Its regulates mitochondrial oxidative phosphorylation, glycolysis, glutamine metabolism, lipid metabolism, and antioxidant defense. Through the regulation of these metabolic processes, p53 maintains the homeostasis of cellular metabolism and redox balance in cells. How this activities contributes to the role of TP53 as a tumor suppressor is currently unknown.
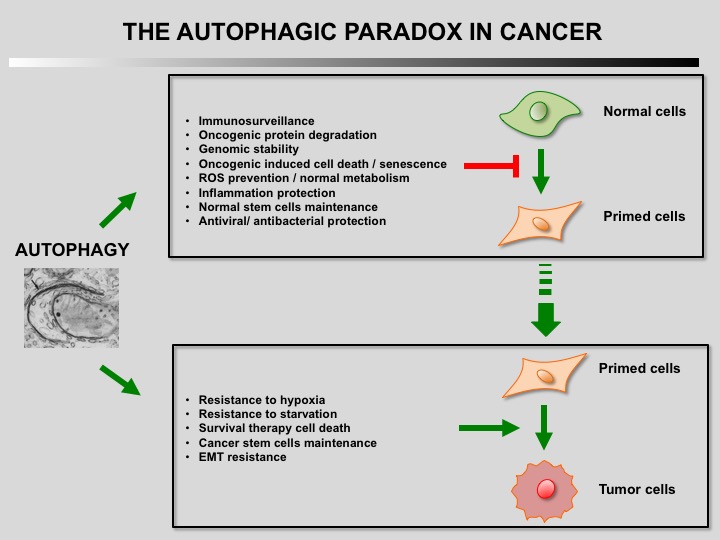
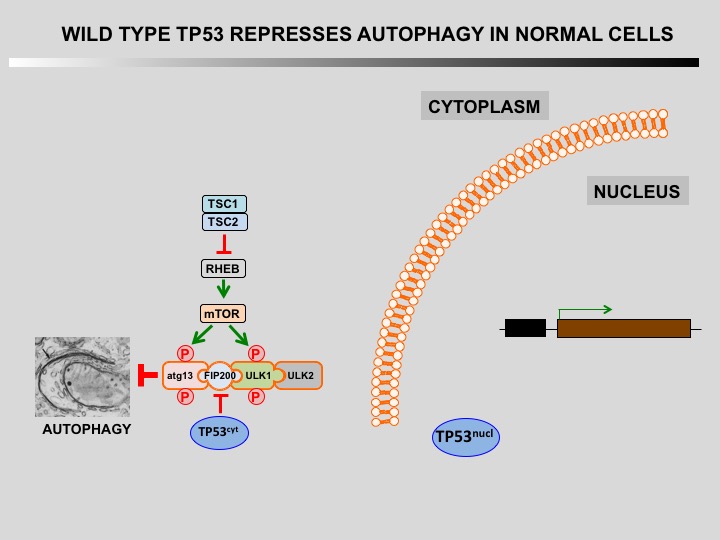
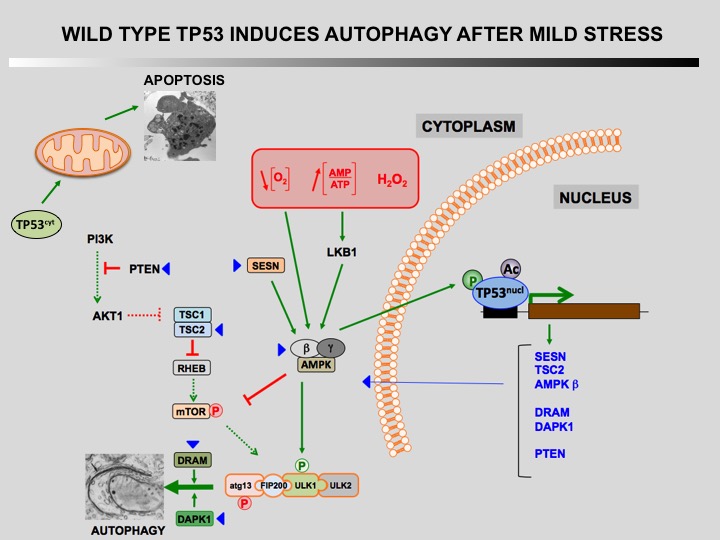
Autophagy is a cellular self-catabolic process in which cytoplasmic constituents are sequestered in double membrane vesicles that fuse with lysosomes where they are degraded. Autophagy is often induced by various forms of mild stress providing a mechanism to maintain cell viability. It is executed at basal levels in every cell and promotes cellular homeostasis and tumor suppression.
Defect of autophagy in preneoplastic cells can promote tumorigenesis leading to a transformed phenotype. Restauration of autophagy is then essential for the survival of tumoral cells.
Bieging KT, Mello SS, Attardi LD (2014) Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer 14: 359-370.
Berkers CR, Maddocks OD, Cheung EC, Mor I, Vousden KH (2013) Metabolic Regulation by p53 Family Members. Cell Metab 18: 617-633.
Beckerman R, Prives C (2010) Transcriptional regulation by p53. Cold Spring Harb Perspect Biol 2: a000935.
Laptenko O, Prives C (2006) Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ 13: 951-961.
Levine AJ, Hu W, Feng Z (2006) The P53 pathway: what questions remain to be explored? Cell Death Differ 13: 1027-1036.
Harris SL, Levine AJ (2005) The p53 pathway: positive and negative feedback loops. Oncogene 24: 2899-2908.
Galluzzi, L., Pietrocola, F., Bravo-San Pedro, J. M., Amaravadi, R. K., Baehrecke, E. H., Cecconi, F., Codogno, P., Debnath, J., Gewirtz, D. A., Karantza, V., Kimmelman, A., Kumar, S., Levine, B., Maiuri, M. C., Martin, S. J., Penninger, J., Piacentini, M., Rubinsztein, D. C., Simon, H. U., Simonsen, A., Thorburn, A. M., Velasco, G., Ryan, K. M., and Kroemer, G. (2015). Autophagy in malignant transformation and cancer progression. EMBO J 34, 856-880.