Analysis p53 mutations heterogeneity in human tumours
The objective of our group is to understand how alterations of the p53 tumor suppressor gene contribute to the heterogeneity of the clinical manifestations of human cancer using a multidisciplinary program combining clinical, in silico and biological analyses
More than 50% of human tumors carry TP53 gene mutations and, consequently, more than 45,000 somatic and germline mutations have been collected in the UMD TP53 database developed in our laboratory (http://p53.fr). Analyses of these mutations have been invaluable for furthering our knowledge on the structure–function relationships within the TP53 protein and the high degree of heterogeneity of the various TP53 mutants in human cancer. Using the evolution and content of the TP53 LSDB as a paradigm, it has been possible to construct a model of gene mutation analysis covering initial descriptions, the accumulation and organization of knowledge in databases, and the use of this knowledge in clinical practice. It has also been possible to formulate several assumptions on the future of LSDBs and how centralized databases could change the accessibility of data, with interfaces optimized for different types of users and adapted to the specificity of each region of the genome, coding or noncoding, associated with tumor development
TP53 mutations are heterogeneous among the various types of cancers. This can be linked to either the specific mutagenic pattern associated with the various types of cancer or a specific selection for TP53 in a given cancer. Developing novel in vitro and in vivo models for these specific variants should provide novel insight into the oncogenic functions of TP53 mutants.
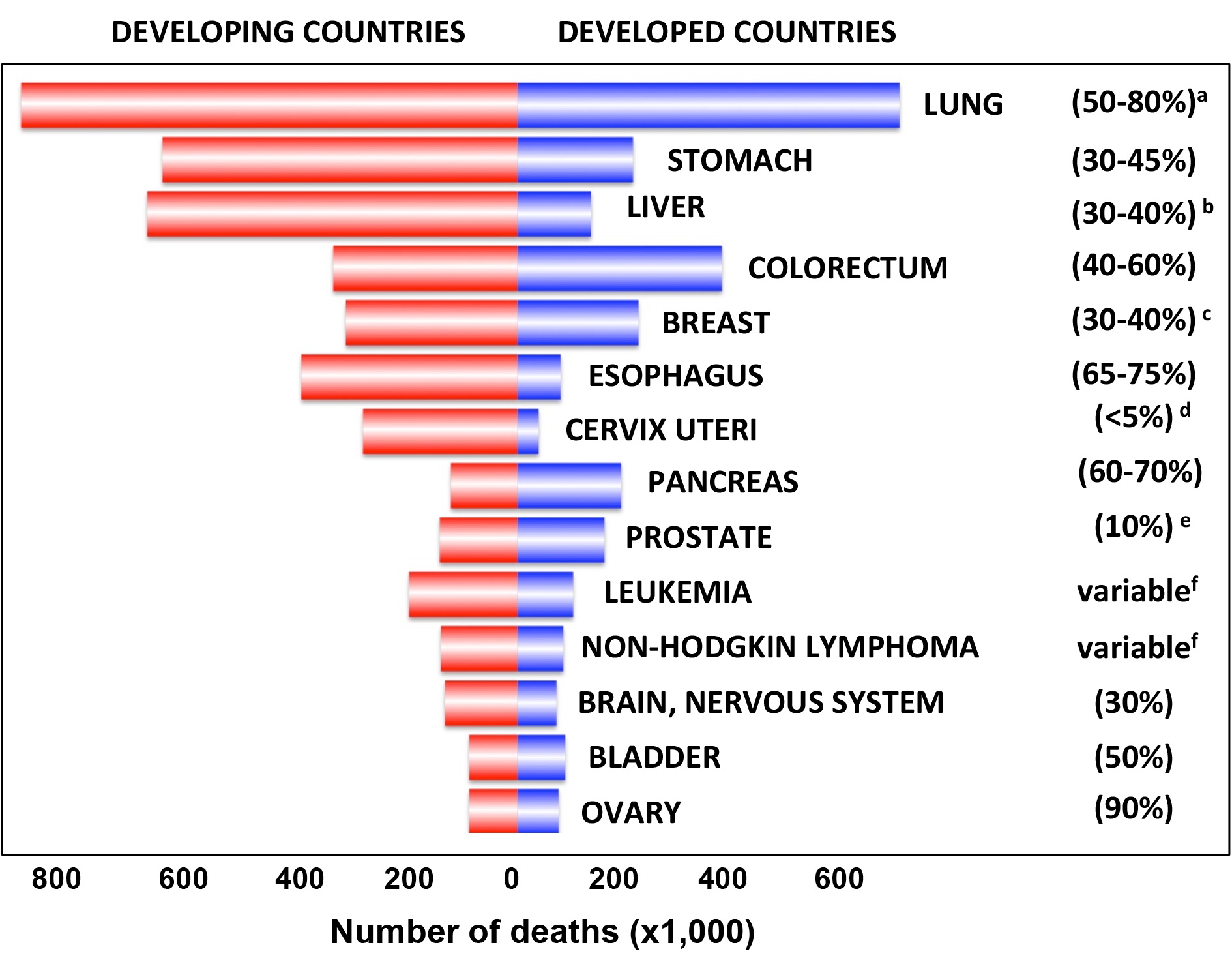
|
Frequency of cancer deaths worldwide and relationship to the frequency of TP53 mutations. Cancer death numbers from GLOBOCAN 2008 (http://globocan.iarc.fr: TP53 mutation data from the UMD TP53 database 2017 a) The frequency of TP53 mutations in lung cancer varies among the diverse subtypes; |
Recent publications
Soussi T & Kroemer G (2017) TP53 and 53BP1 Reunited. Trends Cell Biol, 27: 311–313
Soussi T, Taschner PE & Samuels Y (2017) Synonymous Somatic Variants in Human Cancer Are Not Infamous: A Plea for Full Disclosure in Databases and Publications. Hum Mutat, 38: 339–342
Leroy B, Ballinger ML, Baran-Marszak F, Bond GL, Braithwaite A, Concin N, Donehower LA, El-Deiry WS, Fenaux P, Gaidano G, Langerød A, Hellstrom-Lindberg E, Iggo R, Lehmann-Che J, Mai PL, Malkin D, Moll UM, Myers JN, Nichols KE, Pospisilova S, Ashton-Prolla P, Rossi D, Savage SA, Strong LC, Tonin PN, Zeillinger R, Zenz T, Fraumeni JF, Taschner PE, Hainaut P & Soussi T (2017) Recommended Guidelines for Validation, Quality Control, and Reporting of TP53 Variants in Clinical Practice. Cancer Res, 6: 1250–1260
Lazarian G, Tausch E, Eclache V, Sebaa A, Bianchi V, Letestu R, Collon JF, Lefebvre V, Gardano L, Varin-Blank N, Soussi T, Stilgenbauer S, Cymbalista F & Baran-Marszak F (2016) TP53 mutations are early events in chronic lymphocytic leukemia disease progression and precede evolution to complex karyotypes. Int J Cancer, 139: 1759–1763
Lodé L, Cymbalista F & Soussi T (2016) Genetic profiling of CLL: a ‘TP53 addict’ perspective. Cell Death Dis, 7: e2042
Soussi T & Wiman KG (2015) TP53: an oncogene in disguise. Cell Death Differ, 22: 1239–1249
Soussi, T. (2014). Locus-Specific Databases in Cancer: What Future in a Post-Genomic Era? The TP53 LSDB Paradigm. Hum Mutat 35, 643-653.
Leroy, B., Anderson, M., and Soussi, T. (2014). TP53 Mutations in Human Cancer: Database Reassessment and Prospects for the Next Decade. Hum Mutat 35, 672-688.
Soussi, T. (2014). The TP53 gene network in a postgenomic era. Hum Mutat 35, 641-642.
Soussi, T., Leroy, B., and Taschner, P. E. (2014). Recommendations for Analyzing and Reporting TP53 Gene Variants in the High-Throughput Sequencing Era. Hum Mutat 35, 766-778.
Leroy, B., Girard, L., Hollestelle, A., Minna, J. D., Gazdar, A. F., and Soussi, T. (2014). Analysis of TP53 Mutation Status in Human Cancer Cell Lines: A Reassessment. Hum Mutat 35, 756-765.
Edlund K, Larsson O, Ameur A, Bunikis I, Gyllensten U, Leroy B, Sundstrom M, Micke P, Botling J, and Soussi T. 2012. Data-driven unbiased curation of the TP53 tumor suppressor gene mutation database and validation by ultra deep sequencing of human tumors. Proc Natl Acad Sci U S A 109: 9551-9556.
