Current release: October 2017 (2017_R2): 80,400 tumors, 6,870 diffreent TP53 variants
The 2017 release of the UMD_TP53 database is a major update that takes into account novel issues related to the structure of the TP53 gene, TP53 isoforms, mutation nomenclature and the release of the sequence of more than 5,000 tumor samples from 12 cancer types. This database includes also novel mutations that target specifically TP53 isoforms beta and gamma.
Two different outpouts from the database are currently available:
The UMD TP53 mutation database includes the TP53 status of more than 80,400 tumors, individuals with germline mutations and cell lines.
The UMD TP53 variant database includes a full description of the 6,870 TP53 variants found in the database with 70 novel features associated with each TP53 variant.
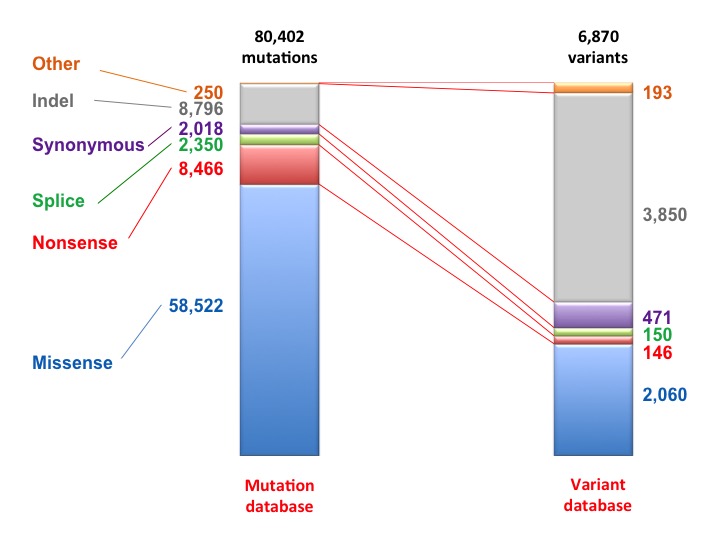
Relation between TP53 variants and individual cases in the UMD TP53 mutation database. Although, 3,850 different indels have been indentified (52% of all TP53 variants), they are only found in 10% of patients. Non synomymous TP53 variants (2,060 variants) are the most frequent alteration found in the TP53 gene (49,950 cases).
The mutation database
The UMD_TP53 database is the only mutation database that provides a curated set of TP53 mutations . Each publication is associated with a confidence index that allows the user to choose to work with a specific dataset (see the curation page and the read me file for more info).
Novelties in the 2017 release of the database
- Mutation description has been updated using Hg18/Build36, Hg19/Build37as well as the novel GRCh38/hg38 assembly of the human genome
- Mutation position has been checked using Mutalyser (https://www.mutalyzer.nl/) using recommendations from the Human Genome Variation Society (http://www.hgvs.org/mutnomen/)
- Mutations for each TP53 isoform are available using the LRG coordinates
- Mutations from the TCGA and ICGC are included
- Statistical analysis to infer the quality of each publication and to identify artifactual mutations has been improved
- Predictive pathogenicity is available for each TP53 variant.
- Each cancer type is now associated with its ICD-10 code (International Statistical Classification of Diseases and Related Health Problems (ICD), a medical classification used by the World Health Organization (WHO)
- Data from dbSNFP including prediction data from multiple algorithms
A full description of this database is available in the "Read Me cases" document.
The variant database
Each variant is associated with more than 70 features such as localization in the various TP53 isoforms, remaining activity, frequency in the database, distribution in various cancer types or prediction of pathogenicity using multiple algorithms (SIFT, Polyphen, CONDEL, PROTEAN or mut_assessor).
This table can be used to assess the pathogenicity of each TP53 variants.
A full description of this variant database is available in the "Read Me variant" document.
Last references related to this issue of the TP53 database
Leroy, B., Anderson, M., and Soussi, T. (2014). TP53 mutations in human cancer: database reassessment and prospects for the next decade. Hum Mutat 35, 672-688.
Soussi T, Leroy B, Taschner PE (2014) Recommendations for analyzing and reporting TP53 gene variants in the high-throughput sequencing era. Hum Mutat 35: 766-778.
Leroy, B., Fournier, J. L., Ishioka, C., Monti, P., Inga, A., Fronza, G., and Soussi, T. (2013). The TP53 website: an integrative resource centre for the TP53 mutation database and TP53 mutant analysis. Nucleic Acids Res 41, D962-D969.
Leroy, B., M. L. Ballinger, F. Baran-Marszak, G. L. Bond, A. Braithwaite, N. Concin, L. A. Donehower, W. S. El-Deiry, P. Fenaux, G. Gaidano, A. Langerød, E. Hellstrom-Lindberg, R. Iggo, J. Lehmann-Che, P. L. Mai, D. Malkin, U. M. Moll, J. N. Myers, K. E. Nichols, S. Pospisilova, P. Ashton-Prolla, D. Rossi, S. A. Savage, L. C. Strong, P. N. Tonin, R. Zeillinger, T. Zenz, J. F. Fraumeni, P. E. Taschner, P. Hainaut, and T. Soussi. 2017. Recommended Guidelines for Validation, Quality Control, and Reporting of TP53 Variants in Clinical Practice. Cancer Res 6: 1250-1260.
